Interfacial and Deep Cholesterol Binding Sites on Mammalian Membrane Proteins
Notes
SITE STILL IN PREPARATION.
Click on pictures for a larger image
 |
 |
 |
 |
|
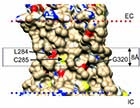
(A) Surface of β2-adrenergic receptor |
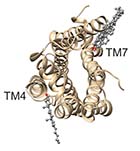
(B) CGMD simulation of β2-adrenergic receptor |
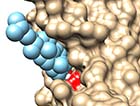
(C) Docking to the β2-adrenergic receptor |
(D) Docking to Gly320 in the β2-adrenergic receptor |
The site provides information on binding of cholesterol to membrane proteins derived from docking studies using AutoDock Vina.
Two types of binding site are described:
1. Deep cholesterol binding sites, located deep within the hydrophobic core of the membrane, as described in:
S.Genheden, J.W.Essex, and A.G.Lee, 'G protein coupled receptor interactions with cholesterol deep in the membrane', Biochim. Biophys. Acta, 1859, 268-281 (2017). https://doi.org/10.1016/j.bbamem.2016.12.001
A.G.Lee, 'A database of predicted binding sites for cholesterol on membrane proteins, deep in the membrane'. Biophys. J. 115, 522-532 (2018). doi: https://doi.org/10.1016/j.bpj.2018.06.022
2. Interfacial cholesterol binding sites at the membrane interfaces (ie ‘normal’ cholesterol binding sites), as described in
A.G.Lee, 'Interfacial binding sites for cholesterol on G protein coupled receptors'. Biophys J., 116, 1586-1597 (2019). doi: https://doi.org/10.1016/j.bpj.2019.03.025
A.G.Lee, 'Interfacial binding sites for cholesterol on TRP ion channels'. Biophys. J., 117, 2020-2033 (2019). doi: https://doi.org/10.1016/j.bpj.2019.10.011
A.G.Lee, 'Interfacial binding sites for cholesterol on Kir, Kv, K2P, and related Potassium Channels'. Biophys. J., 119, 35-47 (2020). doi: https://doi.org/10.1016/j.bpj.2020.05.028
A.G.Lee, 'Interfacial binding sites for cholesterol on GABAA receptors and competition with neurosteroids'. Biophys. J., 120, 2710-2722 (2021). doi: https://doi.org/10.1016/j.bpj.2021.05.009
A.G.Lee, 'The role of cholesterol binding in the control of cholesterol by the Scap-Insig system.' European Biophysics Journal, in press. doi: https://doi.org/10.1007/s00249-022-01606-z9
The menu on the right provides tables of data for bound cholesterols, together with PDB files for download that include protein coordinates and coordinates for bound cholesterols. For interfacial sites, PDB files are provided separately for cholesterols bound on the extracellular (EC) and intracellular sides (IC) of the membrane.
Viewing the downloaded PDB files:
1. Interfacial sites. The PDB files should be viewable in all common viewers.
2. Deep binding sites. These files have a format similar to that adopted for multiple NMR structures, with the first ‘model’ being the protein structure and a series of extra models, each of which contains one of the docked cholesterol modes. The files open directly in Chimera and in Discovery Studio Visualizer and also in Pymol as long as the ‘set all states, on’ option has been set by typing ‘set all states, on’ in the command line. The files cannot be opened directly in VMD as VMD does not support multi-model files. However, a file splitter program such as multiple_model_PDB_file_splitter.py (free on the web) can be used to generate a set of PDB files from the original file, one containing the protein structure, the others each containing one docked cholesterol mode. These PDB files can then be loaded into VMD in the usual way.